Immediately after the infection of a cell in the throat or lungs, the SARS-CoV-2 virus works very hard to replicate, using the human cell’s metabolic pathways to produce its proteins and make sure that its genetic material (the RNA genome) is copied. The RNA genome is then packaged very compactly into new virus particles that are released from the cell to infect more cells.
One viral protein, called the nucleocapsid protein (N), is particularly important for rapid and efficient replication. It wraps around the RNA genome in the virus and ensures that the very long RNA is tightly coiled up. When it penetrates the cell, N detaches itself from the RNA genome and assumes a whole range of functions during viral replication: When the RNA is translated into viral proteins, N protects the RNA from being destroyed by the cell’s antiviral defense mechanism (“RNA interference“). N also contributes directly to the transcription of RNA into viral proteins, and finally it collects the replicated viral RNA in the cell and coils it up so that new viral particles can form.
Like a Swiss army knife, N has several tools at its disposal for all these functions: Firstly, N must be able to distinguish between cellular and viral RNA and to coil up the latter in a spiral shape. That is why N can bind viral RNA in a relatively non-specific manner. To steer the transcription of viral RNA into viral proteins (translation), for example, N must, however, equally be able to recognize specific positions on the viral RNA, called RNA motifs.
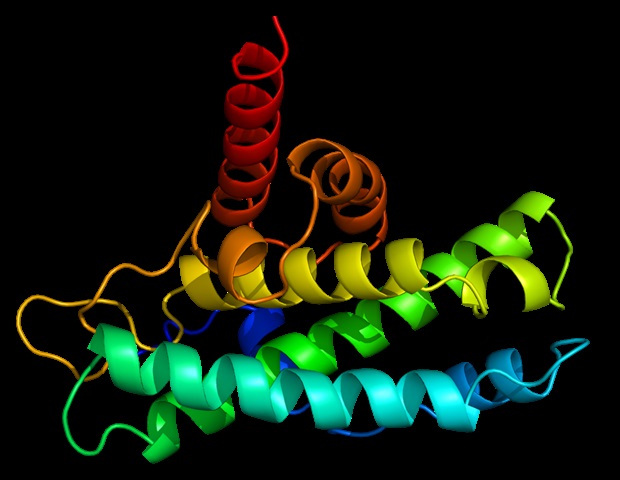
Researchers led by Dr. Sophie Korn and Dr. Andreas Schlundt from the Institute for Molecular Biosciences and the Center for Biomolecular Magnetic Resonance (BMRZ) at Goethe University Frankfurt have now shed light on exactly how this specific binding through one of N’s tools, known as the N-terminal domain (NTD), works. Their results build on preliminary studies by the COVID19-NMR consortium established in Frankfurt during the pandemic. In the work now presented, Korn and her colleagues used nuclear magnetic resonance (NMR) spectroscopy, in which the atoms of the NTD tool and of the bound RNA are exposed to a strong magnetic field and in this way reveal something about their spatial arrangement during binding. In addition, a special X-ray technique (small-angle X-ray scattering, SAXS) delivered precise information about the stability of the newly formed molecular complexes.
The result: Both the sequence of the RNA building blocks (bases) and the spatial folding of the RNA are important for binding, whereby the positively charged part of the NTD binds the negatively charged RNA in a very unspecific way. Several “fingers” of the NTD then explore the RNA in search of motifs that the NTD can use to bind more stably. What attracted the researchers’ attention was that the NTD prefers motifs which are present in lung cells at body temperature in a specific spatial folding that is lost when the temperature increases by just a few degrees. This not only identifies them as their own target motifs but also binds them much more tightly, which could lead to the NTD exercising new functions.
Although our data are only a first step, they suggest that the virus could switch between replication and packaging into new virus particles in this way: At normal body temperature, the cell predominantly produces building blocks for new viruses. If we develop a fever in the course of the infection because our immune system recognizes and fights the virus, the virus might switch to replication as a direct result and ensure that the viral RNA is packaged more and released in the form of new virus particles. It is the viral RNA motifs themselves that provide the switch, which is then triggered by the human defense system.”
Dr. Sophie Korn, Goethe University Frankfurt
Andreas Schlundt is convinced: “With the combination of NMR spectroscopy and SAXS, we have established an experimental method that we can use to quickly assess which binding partners N prefers, and this can probably be transposed to other viral proteins. This will be useful both in the study of emerging viruses and viral variants as well as in the development of antiviral drugs that systematically disable the virus, minimizing side effects in the process.”
Source:
Goethe University Frankfurt
Journal reference:
Korn, S. M., et al. (2023). The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5’-genomic RNA elements. Nature Communications. doi.org/10.1038/s41467-023-38882-y