In a recent early-release article published in the journal Emerging Infectious Diseases, Centers for Disease Control and Prevention researchers investigate and describe the unusual mortality of a cohort of gray (Halichoerus grypus) and harbor (Phoca vitulina) seals infected by a highly pathogenic strain (clade 2.3.4.4b) of the avian influenza (HPAI) A (H5N1) virus. The mortality event was identified in the St. Lawrence Estuary, Quebec, Canada, and comprised 15 dead seals, which necropsy confirmed succumbed to the viral infection.
Research: Outbreak of Highly Pathogenic Avian Influenza A(H5N1) Virus in Seals, St. Lawrence Estuary, Quebec, Canada. Image Credit: davemhuntphotography / Shutterstock
The researchers conducted detailed postmortem examinations of the seal carcasses and, subsequently, molecular sequencing, thereby revealing that the phylogenetic origins and subtypes of the H5N1 lineages were either exclusively Eurasian or a combination (reassortment) of Eurasian with North American genome constellations. The concurrent presence of a large number of HPAI H5N1 infected seabird carcasses at seal haul-out sites suggests that this event represents a host jump event from birds to seals, raising concerns about the potential establishment of a marine mammalian viral reservoir for this deadly disease and worse, the epidemiological potential for zoonotic spillover to humans and other mammalian taxa.
A brief history of avian influenza in marine mammals
H5N1 is a subtype of the influenza A virus (IAV) that frequently infects birds (both wild and farmed populations) and has recently been found to spill over to cows and other animals living close to these infected birds. First discovered in farmed poultry in Southern China in 1996, the virus is a fast-evolving pathogen that has since been observed to sporadically infect marine mammals, most commonly pinnipeds such as Phoca vitulina (harbor seals) and Halichoerus grypus (gray seals), in the United States and Europe.
Even though mammalian infections, particularly with High-Pathogenic Avian Influenza (HPAI) virus strains, are rare, a growing body of literature suggests an increase in the disease’s prevalence. This highlights the need for preventive measures to restrict transmission, thereby preventing a potential future human epidemic. Research aimed at characterizing circulating viral strains suggests an avian-variant origin that has since mutated, allowing it to cross-species transmit to mammalian hosts from wild aquatic birds, some of which carry the disease across vast swaths of geography during their annual winter migrations.
“Harbor seals seem to be particularly susceptible to IAV infections, and factors such as close contact with wild birds and mammalian adaptations of the virus subtypes have been suggested as drivers in establishing a potential reservoir of IAV in marine mammals.”
The latest identified HPAI H5N1 clade has been named ‘2.3.4.4b A/goose/Guangdong/1/1996 (Gs/GD)’ with its first confirmed North American Atlantic coast victim revealed to be a will gull carcass found in November 2021 in eastern Canada. Alarmingly, following its discovery, the virus has been observed to rapidly spread across North America, even reassorting with native American IAV strains, increasing the cross infectivity of the latter and causing unprecedented mortality in both avian and wild terrestrial mammalian hosts. Notably, the summer of 2022 saw widespread harbor and gray seal mortality across eastern Quebec, Canada, and Maine, USA.
About the study
In the present study, researchers report the identification of H5N1 in marine mammals in the St. Lawrence Estuary, Quebec, Canada. Stranding data for the study was obtained from the Quebec Marine Mammal Emergency Response Network between April 1 and September 30, 2022. The researchers conducted a detailed postmortem (histopathological) examination of 27 frozen seal carcasses, 15 of which (55.56%) were observed to have been fatally infected by HPAI H5N1. Nasal and rectal swab samples from the 15 thawed carcasses submitted for necropsy and six stranded carcasses from a field-based seal landing site were collected for subsequent RNA and phylogenetic analysis.
Phylogenetic analyses were carried out using reverse transcription polymerase chain reaction (RT-PCR) for RNA amplification, followed by next-generation Oxford Nanopore sequencing and genome assembly. A BLAST similarity search was used to identify the genetic relatedness between swab-obtained H5N1 samples and previously characterized HPAI IAV samples from online genome databases collected from wild birds between April and September 2022. Finally, immunohistochemistry assays were carried out to identify HPAI IAV antigens in infected bird and seal carcasses.
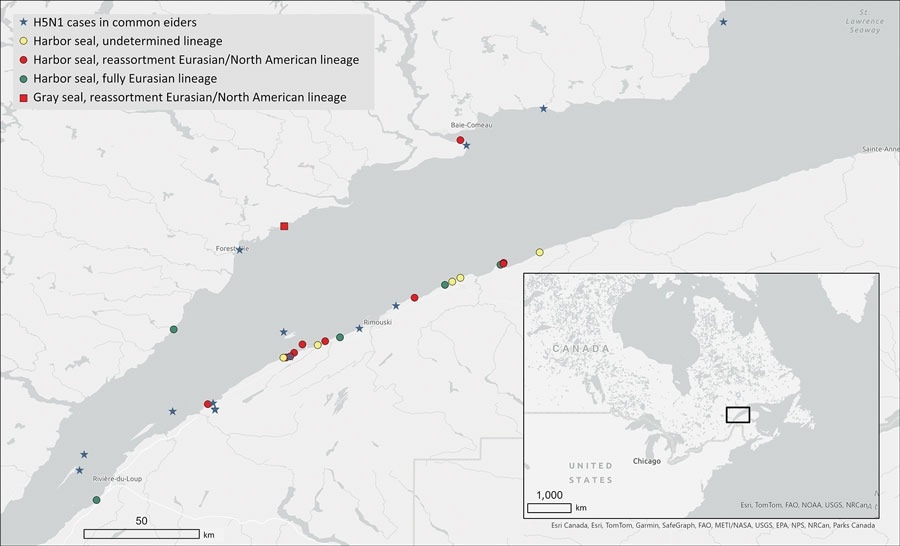
Geographic locations of stranded, dead, or sick seals infected by highly pathogenic avian influenza A(H5N1) virus during the 2022 outbreak in the St. Lawrence Estuary, Quebec, Canada. The locations of harbor seals (Phoca vitulina) gray seals (Halichoerus grypus) and detected H5N1 lineages are marked as are the documented outbreaks in common eider (Somateria mollissima) colonies. Inset shows study location in a map of eastern Canada, and US Midwest and Northeast.
Study findings and conclusions
Descriptive necropsy findings revealed substantial histological changes, particularly fibrinosuppurative alveolitis, multiorgan acute necrotizing inflammation, and meningoencephalitis, the latter of which was identified in all submitted seal carcasses. These histological lesions and associated molecular investigations confirmed that the observed seal mortality was due to HAPI H5 viral infections.
“All the infected adult harbor seals (n = 9) were female, and 6 had evidence of recent parturition (active lactation, asymmetric uterine horns without the presence of a fetus, or both). The infected adult gray seal was male. There were 3 male and 7 female (and 1 nondetermined) infected <1 year old seals. One of the infected seals was found alive with profound lethargy and neurologic signs. In addition, anecdotal observations of weak and dyspneic harbor seals were reported during the outbreak.”
Virology assessments confirmed the presence of IAV H5 RNA and excluded H7 RNA (H7 has previously been suggested as being responsible for some observed marine mammalian mortality). Genomic analyses revealed that all isolates belonged to the Gs/GD lineage H5N1 clade 2.3.4.4b. While five of the 16 included isolates aligned exclusively with Eurasian lineages (suggesting a transoceanic, avian-origin cross infection), the remaining 11 displayed North American elements, highlighting the alarming trend of viral genetic reassortment.
In summary, this study highlights recent viral mutations allowing for the entry and replication of influenza viruses from their ancestral avian hosts into mammalian cells. It sparks concern on two significant fronts – 1. That marine mammals, including seals and whales, may represent a viral reservoir with substantial epidemiological management difficulties, and 2. The continued evolution of H5N1 and other HPAI IAV strains may represent a potentially deadly human epidemic in the future.
Journal reference:
- Lair S, Quesnel L, Signore AV, Delnatte P, Embury-Hyatt C, Nadeau M-S, et al. Outbreak of highly pathogenic avian influenza A(H5N1) virus in seals, St. Lawrence Estuary, Quebec, Canada. Emerg Infect Dis. 2024 Jun, DOI: 10.3201/eid3006.231033, https://wwwnc.cdc.gov/eid/article/30/6/23-1033_article








